Instant sequence search
We’re pleased to announce that we’re launching a new feature on One Codex today: instant sequence search.
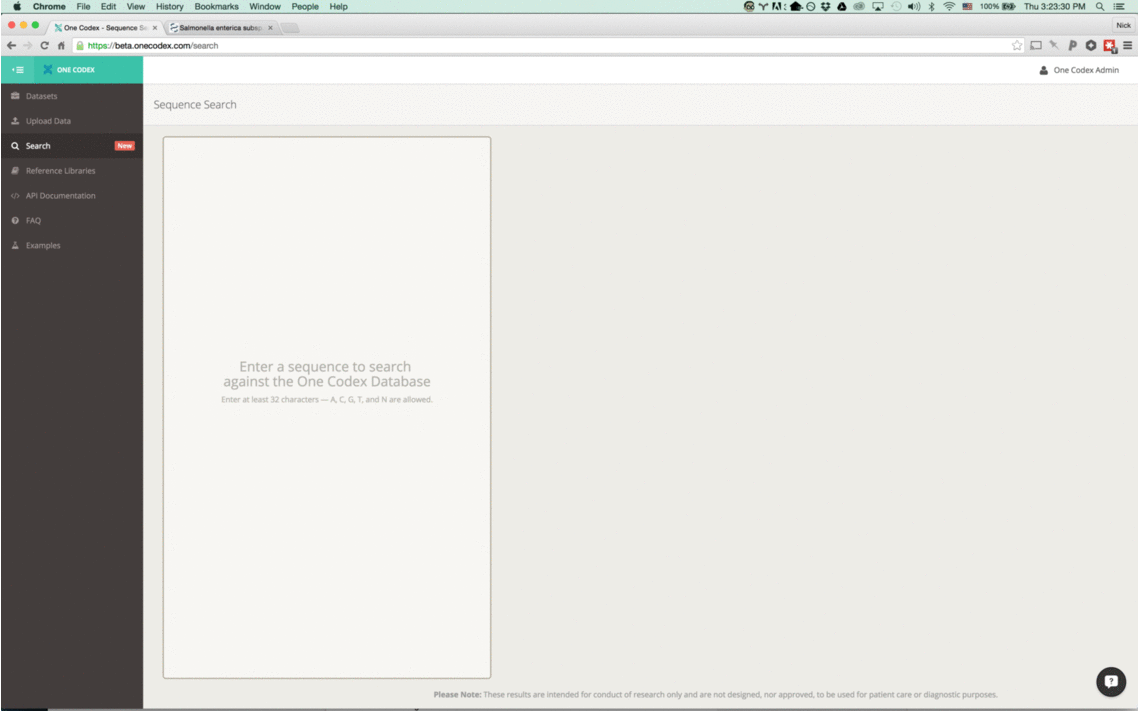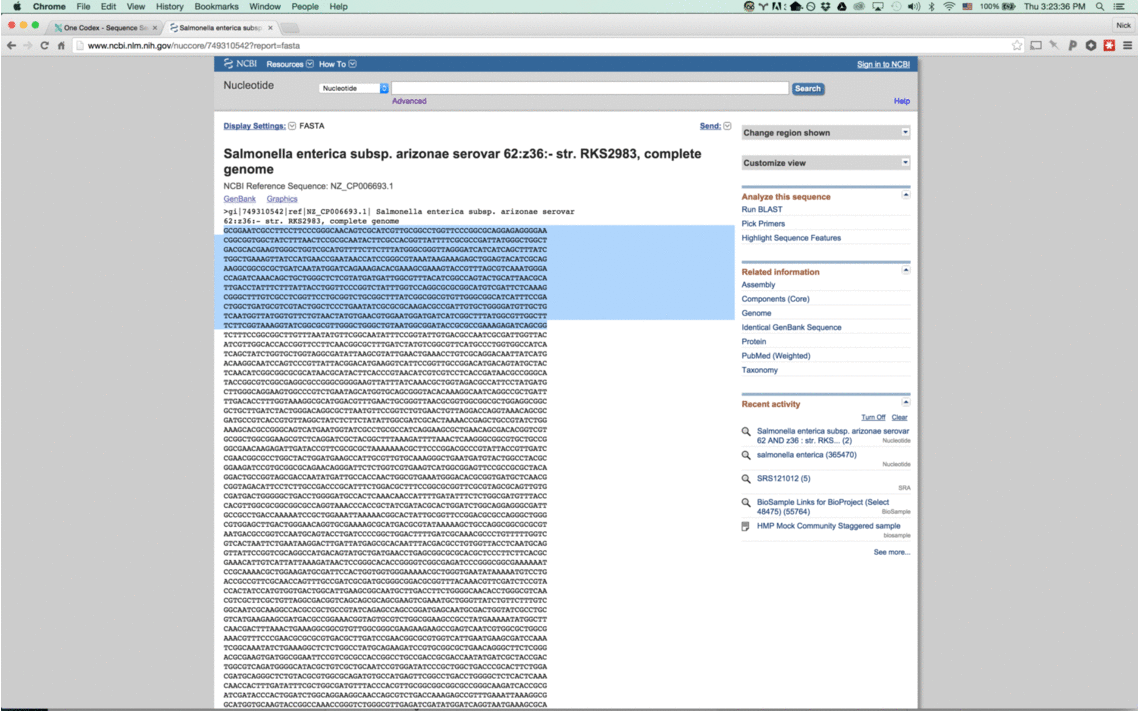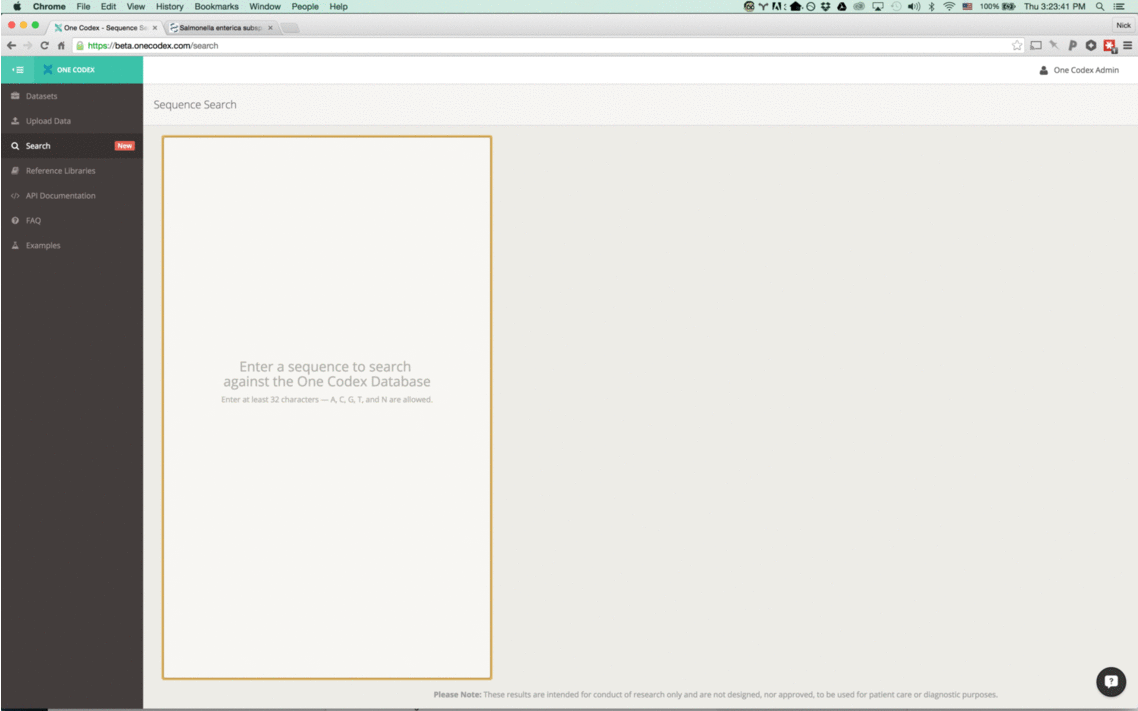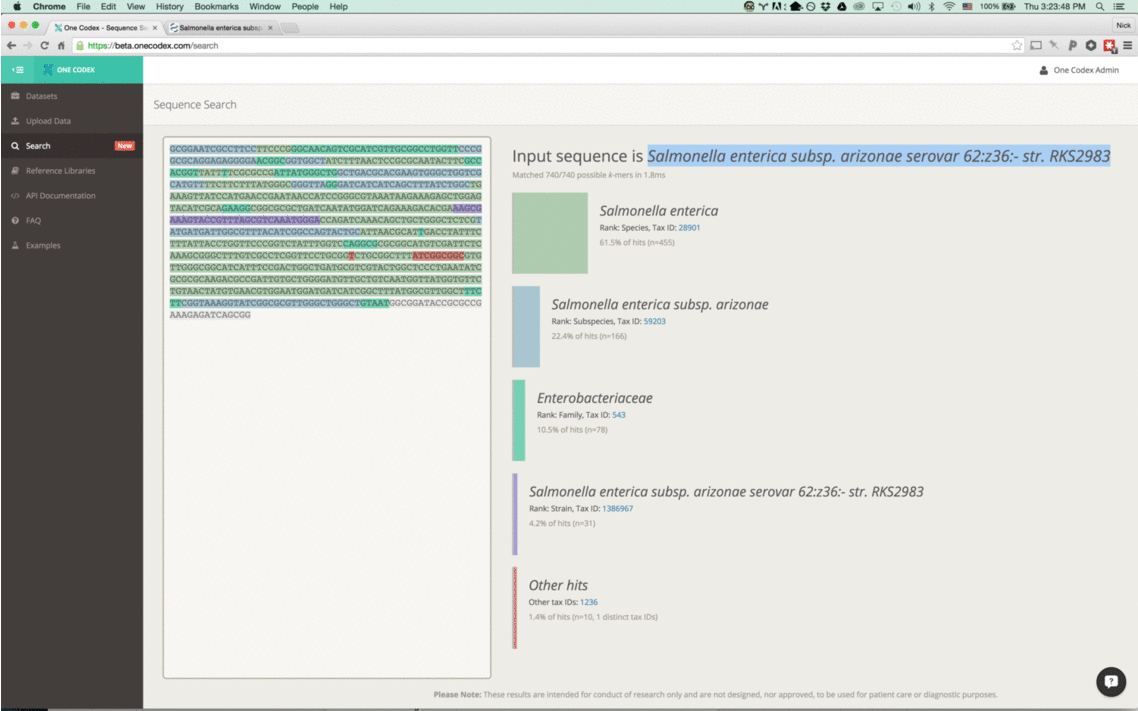
With this new tool, simply enter a nucleotide sequence – whether a read from a FASTQ file or a section of a reference genome – and we’ll instantly compare it against our One Codex database.
How it works
Our instant classifier works by splitting up the input sequence into all overlapping k-mers and then comparing each against our reference database, using a k=31. We then visually highlight the most specific taxonomic value (or lowest common ancestor) for the k-mer in our database. (Note that because the k-mers overlap, we only color the first character of each)