Build a single source of truth for your microbial assets with the brand new Genome Library. Learn more →
Launch your clinical metagenomics workflows on One Codex today
Kickstart your clinical NGS program with the first pathogen-agnostic analysis platform that fits your workflow.
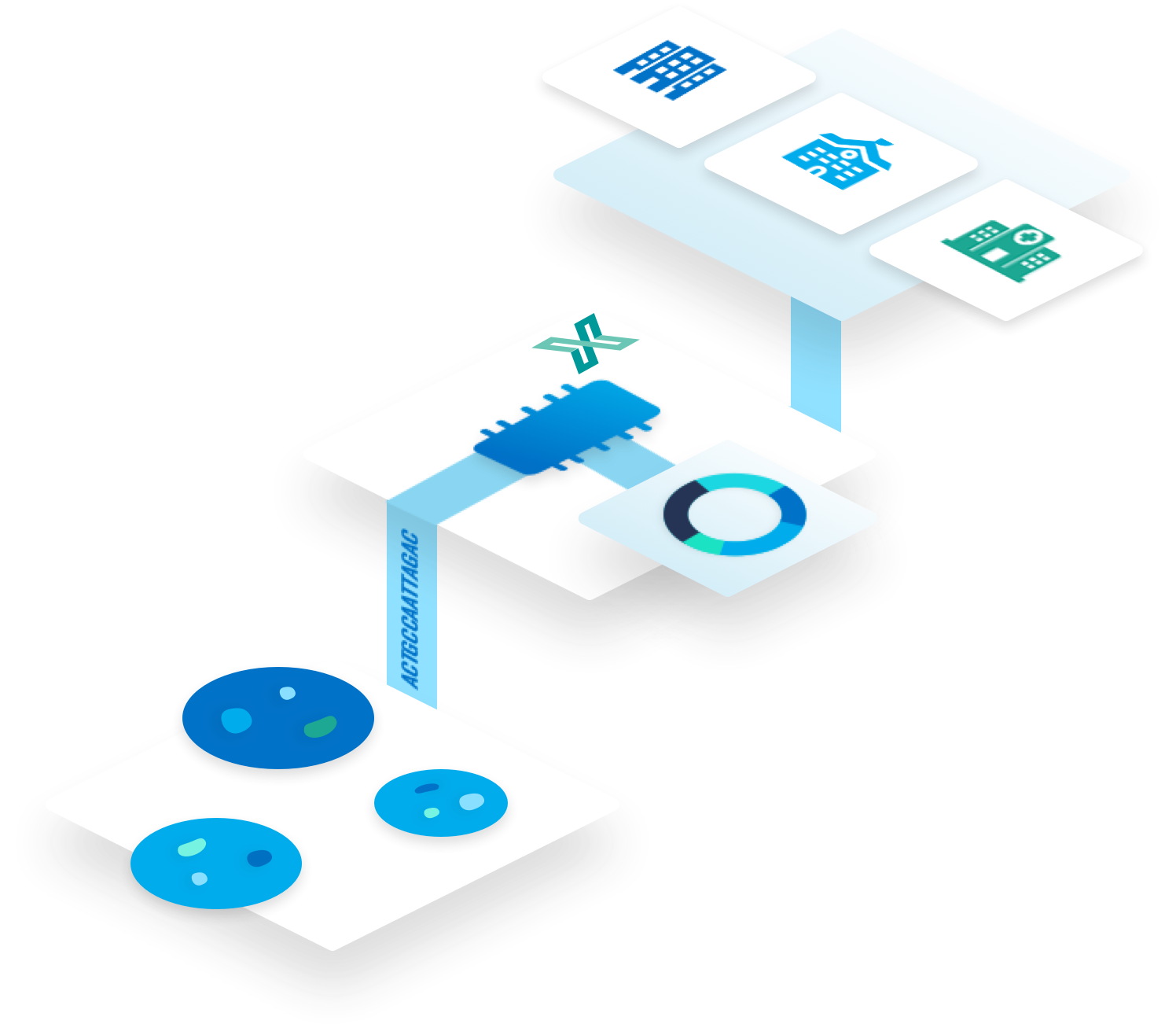
A Cloud-Based Platform for Clinical Metagenomics
Standardized genomics workflows are not currently mainstream in clinical and public health practice.
Variation in methods, expertise, and staff bandwidth leads to inconsistent results and outcomes within institutions and inhibits surveillance across the health system.
One Codex is a cloud based analytical platform specializing in microbial genomics. One Codex is uniquely positioned to deliver clinical metagenomics decision support systems leveraging its comprehensive microbial genomics reference database, its expertise developing in silico detection algorithms, and its flexible reporting and data delivery architecture.
- Identify any microbe from any sample – provide answers closed systems can’t, with the QC information to rule-out or justify detection.
- Fast and automated – from any sequencing instrument to a Lab Director’s report in <20 minutes. Accurate and efficient, without compromises.
- Relevant, actionable molecular information – eliminate noise and better characterize detected microbes with antimicrobial resistance prediction, strain typing, sequence alignments to reference genomes, and more.
- You maintain control – Lab Directors can apply their expertise interpreting molecular and patient data to finalize reports and send informative results back to ordering clinicians.
- Everything in one place – reports, raw data and intermediate analyses are securely stored on the One Codex platform so you can monitor epidemiologic or QC trends.