Comprehensive and accurate bacterial isolate characterization using multi-locus sequence typing
Sequencing, identifying, and characterizing bacterial isolates is a fundamental task in microbiology. One of the earliest, and still widely used, sequence-based methods for isolate characterization is multi-locus sequence typing (MLST). This method unambiguously identifies species isolates using sequences from a handful of an organism’s housekeeping genes [1]; the combinations of alleles at these loci determine the sequence type (ST) of the isolate. Since their inception, these techniques have proven useful, particularly for pathogen surveillance and epidemiological studies [2].
If you’ve ever uploaded whole-genome sequencing (WGS) data of bacterial isolates to your One Codex account, then you’ve already used our MLST analysis! One Codex automatically detects isolate samples, selects the appropriate organism and MLST schemes, performs the analysis, and tags any samples with their resulting STs. All of this happens seamlessly in the background, right after you upload your data.
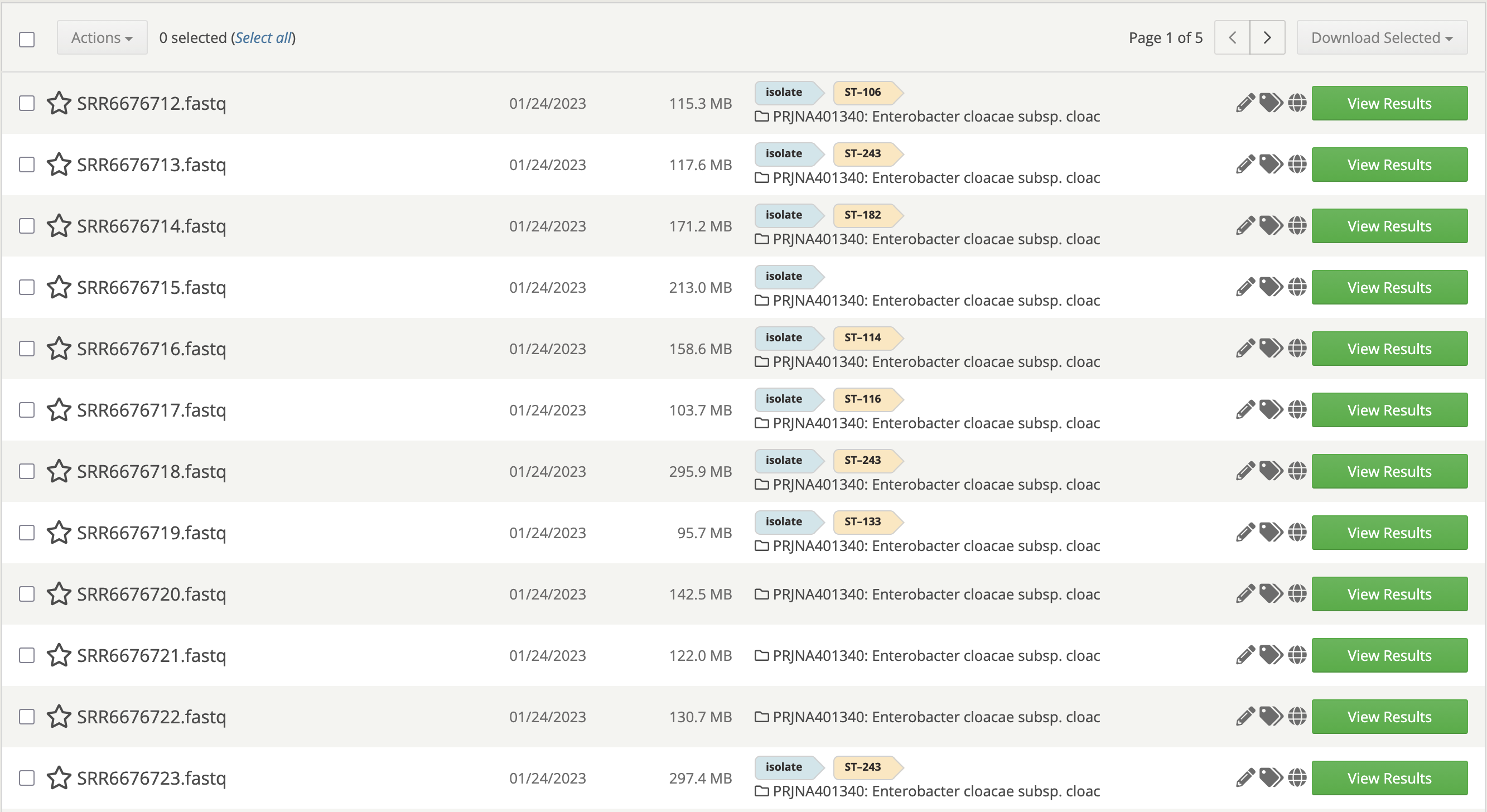
Figure 1. Samples page. Isolate samples are automatically detected and tagged with the appropriate sequence type.
Today, we’d like to bring our MLST analysis to the foreground and share some recent improvements we’ve made. First, we’ve expanded our collection of schemes to support over 100 different species, covering a wide variety of clinically and environmentally relevant bacteria and fungi. These schemes are derived from standardized, open-access resources such as PubMLST [3]. We’ve also added support for long read technologies (e.g. Oxford Nanopore and PacBio), in an ongoing effort to expand long-read support across our platform. Finally, we’ve added scheme information, marker coverage and other useful information to the results page to make it easier to investigate your sequence types.
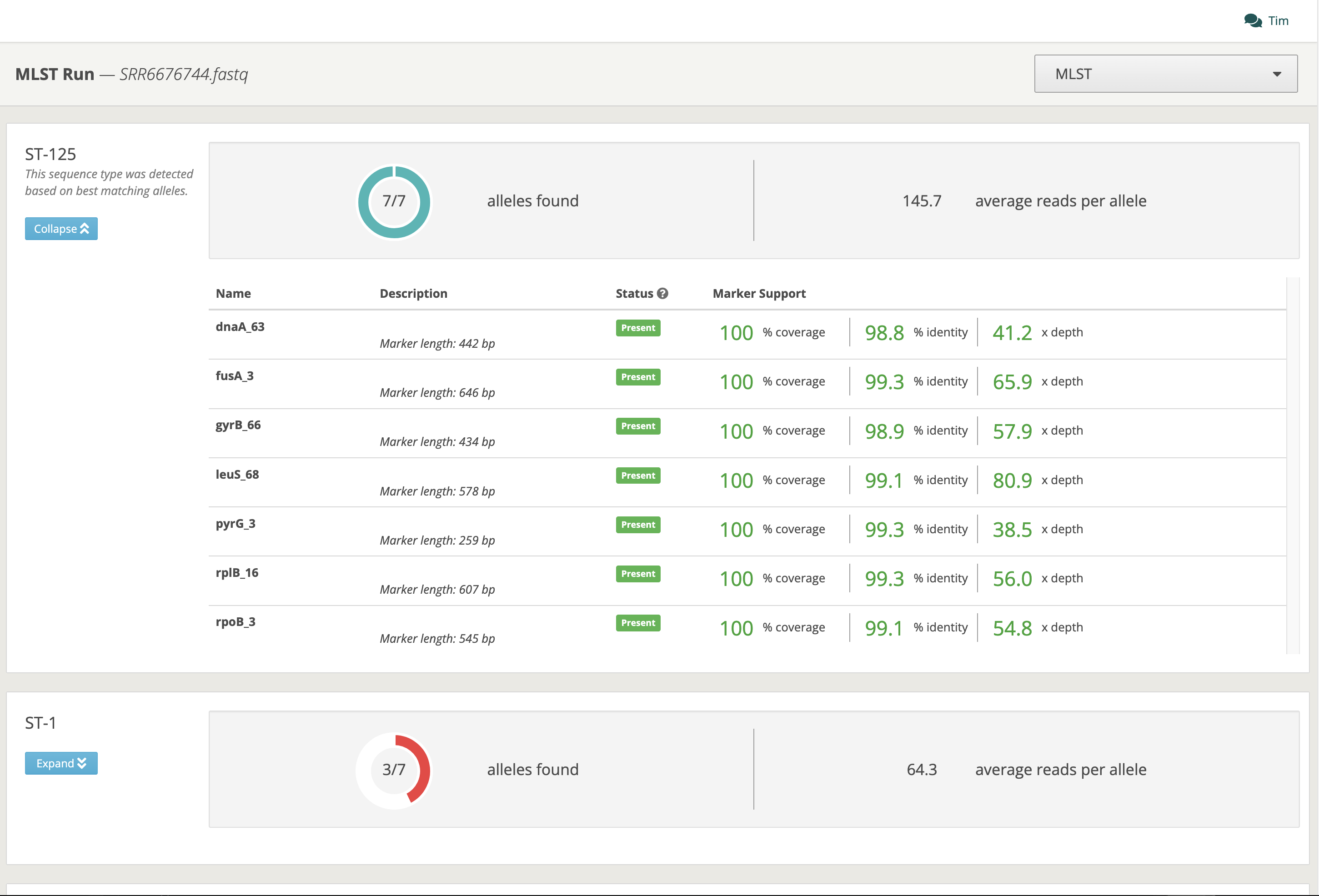
Figure 1. MLST results page for a single sample.
We hope these improvements make it even easier for you to analyze your isolate WGS data. You can get started with our MLST analysis today by uploading isolate samples to your account, or by taking a look at our collection of example isolates, which includes sequencing data from published outbreaks [4]. You can find more information about our MLST analysis in our documentation. As always, we’d love to hear your feedback and suggestions.
References
- Maiden, M. C. et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95, 3140–3145 (1998).
- Urwin, R. & Maiden, M. C. J. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol 11, 479–487 (2003).
- Jolley, K. A. & Maiden, M. C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11, 595 (2010).
- Hawken, S. E. et al. Genomic Investigation of a Putative Endoscope-Associated Carbapenem-Resistant Enterobacter cloacae Outbreak Reveals a Wide Diversity of Circulating Strains and Resistance Mutations. Clin Infect Dis 66, 460–463 (2018).