Introducing Genome Library
The Challenge
Microbial genomics is changing how organizations in research, healthcare, pharma, and agriculture understand their collections of organisms. While many tools exist for addressing the bioinformatics challenges, data organization and accessibility remain major obstacles. Genomic data is often disconnected from metadata and analysis, lacks versioning and provenance data, and is typically found in inscrutable file formats accessible to only a few team members, making it difficult for groups to maximize their investment in microbial genomics.
To tackle these challenges, we’ve created Genome Library—a platform that combines powerful bioinformatics workflows, data management, versioning and organization tools, and a unique real-time sequence search service (more on that below). Together, these tools not only simplify the generation and analysis of genomes, but remove data silos and enable new forms of automation. Using Genome Library, organizations across industries can focus on the biological questions they seek to answer with their genomic information readily at hand.
Our Solution
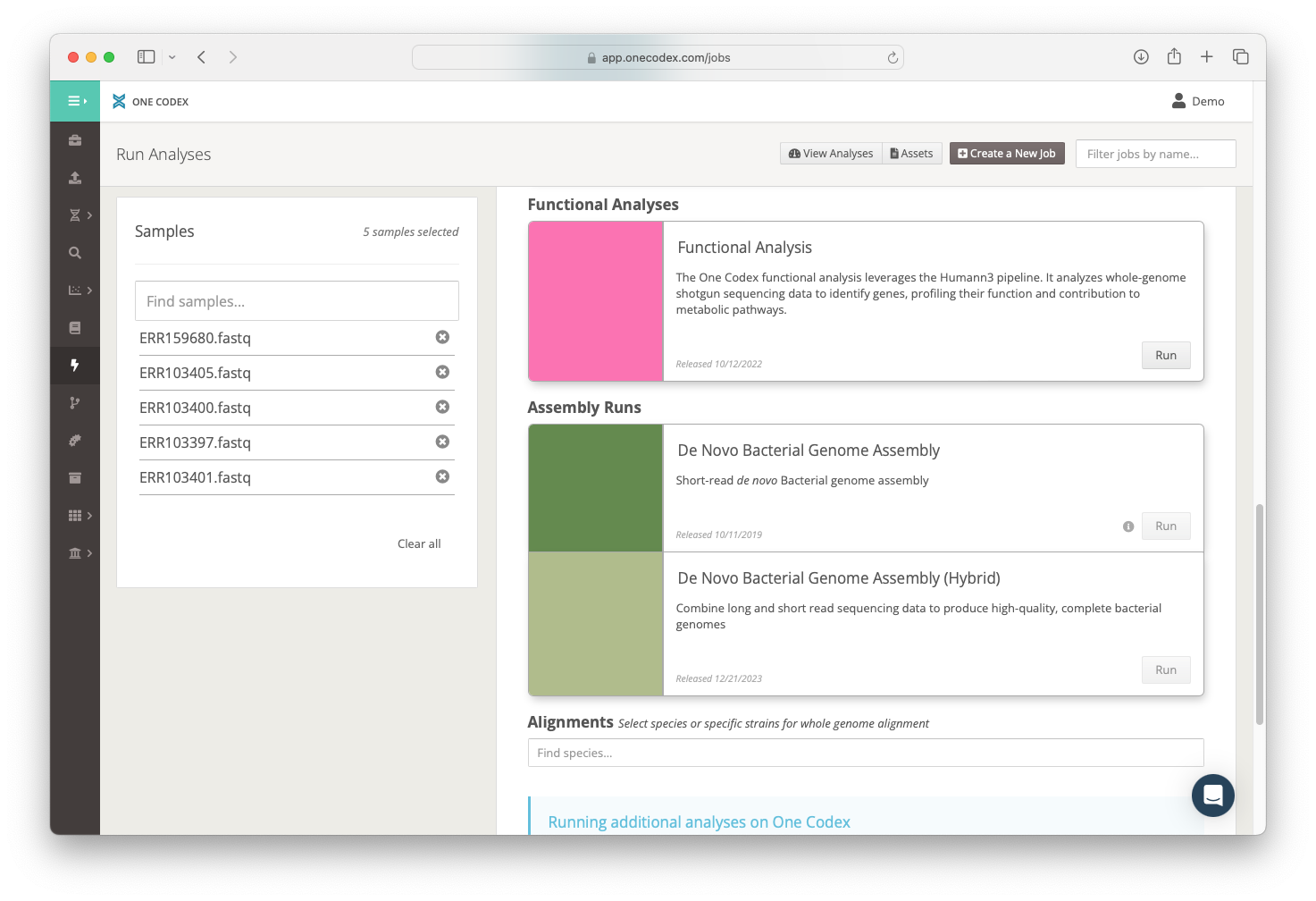
One-Click Assembly Workflows
A major challenge in microbial genomics is obtaining high-quality genome sequences. To make this easier, we are releasing two de novo assembly workflows. Supporting either short read genome assembly, or long-read hybrid assembly (with the addition of either PacBio or Oxford Nanopore reads) with the latter capable of producing fully-closed, reference-grade genomes. All assemblies are automatically annotated and assessed for completeness, contamination and general quality and all workflow versions and input data are linked and easily accessible.
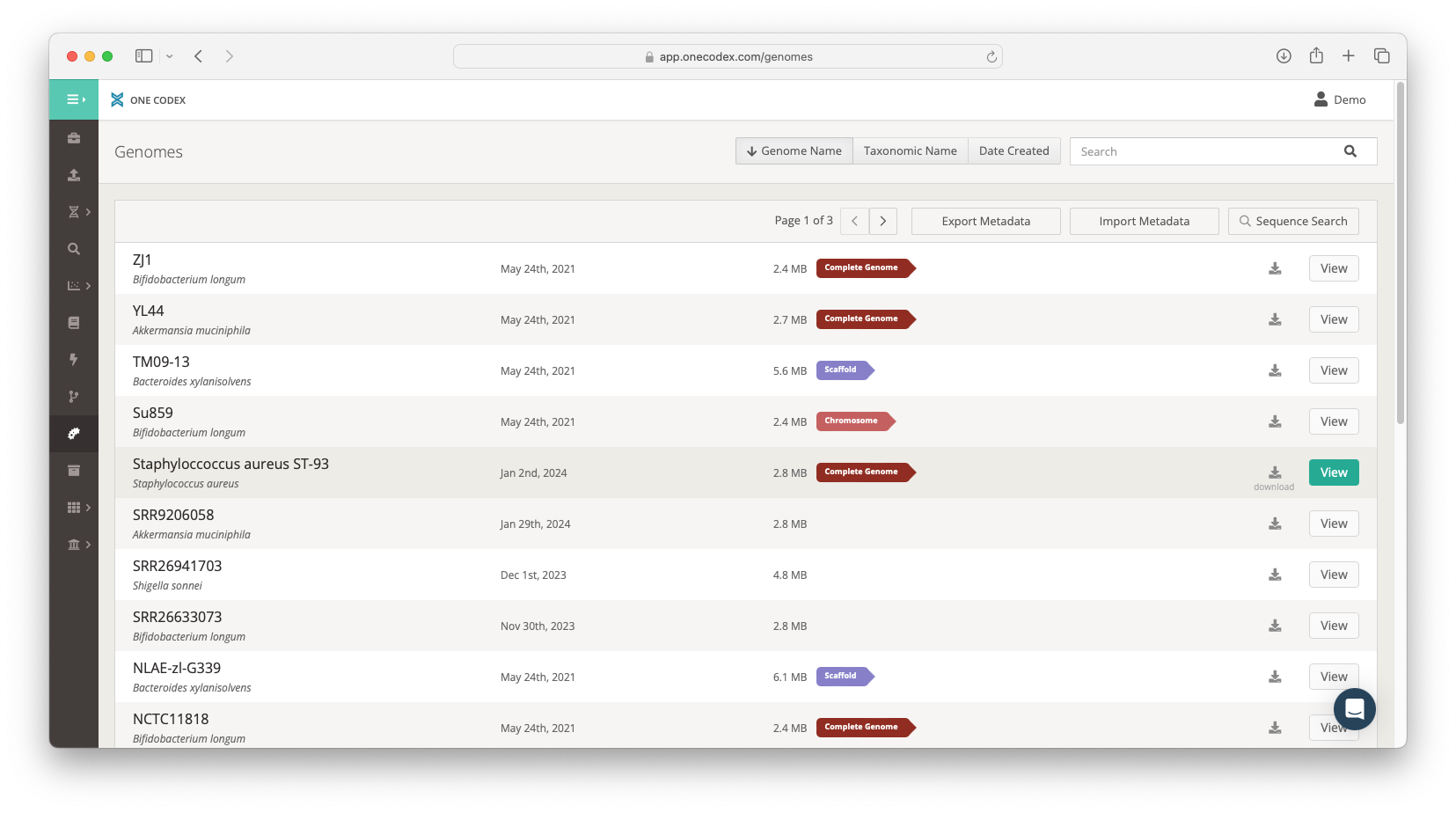
Genome Data Management
Your private library of genome sequences is available on our platform via a user-friendly interface that allows members of your organization to search by taxonomic identity, metadata or sequence content. Members of your organization can also update genome metadata and assembly versions. All data are accessible via our API making it possible to plug into existing data management solutions.
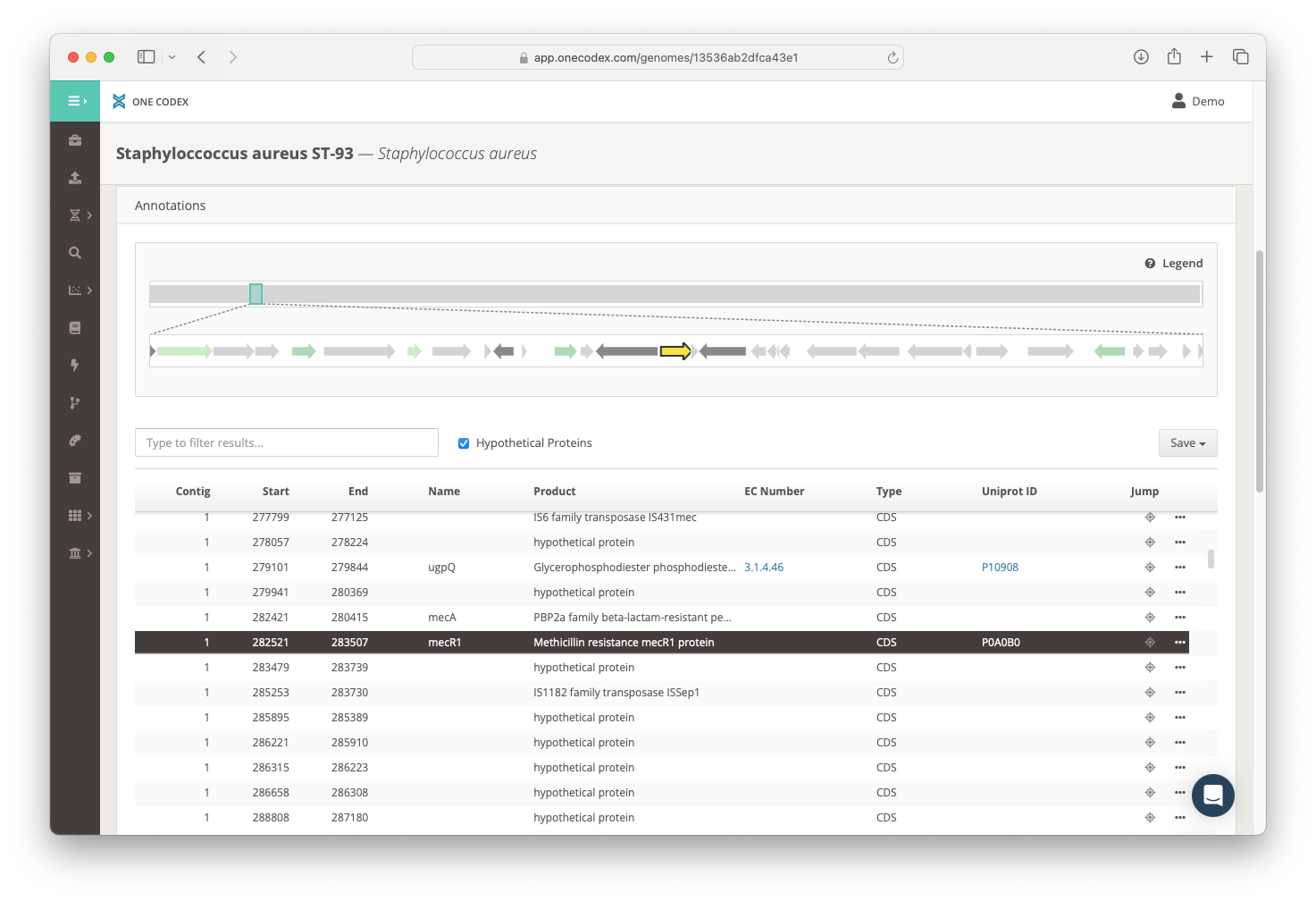
Interactive Genome Browser
We’ve created an interactive genome browser that allows you to quickly search through the gene content of your strains. The browser includes an annotation track to display the physical arrangement of genes within operons. Gene annotations are also searchable and can be exported into more user-friendly formats (or in genbank-format if that’s your thing).

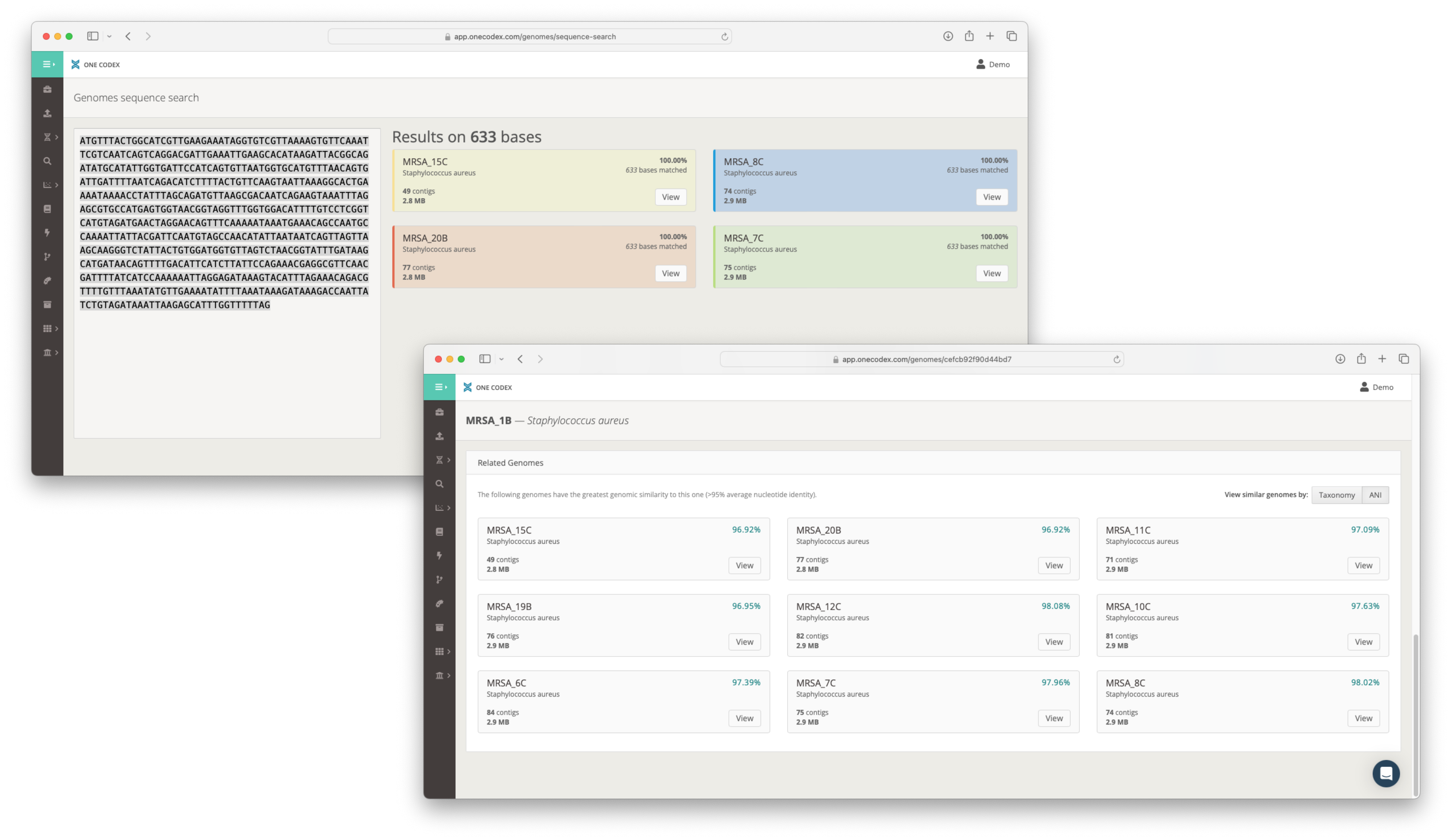
Real-time Sequence Search
Having access to gene annotations is helpful, but not every gene has been characterized and named. To make it possible to find sequences of interest in an annotation-agnostic manner, we’ve built a sequence search engine that allows you to search for similar sequences across your entire genome collection in real-time. This tool automatically stays up to date as your genome collection grows.
Our sequence search engine also allows you to quickly explore the phylogenetic relationships between isolates, instantly showing you which genomes in your collection may be similar to one another. This is especially useful in cases where taxonomic names may be tentative or non-existant.
We’re really excited about Genome Library and all of the potential it has to solve problems encountered across industries. Here are some examples of the types of applications where we think Genome Library fits:
- Strain identification, AMR and pathogen detection results for healthcare, epidemiology and pharmacy
- Verification of identity and integrity of asset organisms
- Tracking lineages of engineered strains and validating gene editing targets
- Mining for microbes that possess interesting pathways while also selecting for genomic diversity
Learn More
Click here to see a live demo or reach out to find out more.