Importing Sequence Read Archive (SRA) Data Into One Codex
We are happy to announce that One Codex now supports importing data from the Sequence Read Archive (SRA), allowing for quick comparison and analysis of publicly available sequencing data and associated metadata.
What is the SRA?
The SRA is a public repository for the storage and retrieval of sequencing data and the associated metadata1. Originally called the Short Read Archive2, and hosting primarily human and mouse DNA3, it has since expanded to host the sequencing data of a greater variety of sample and instrument types. It now hosts long reads, epigenetic, ChIP-seq, single-cell, and metagenomic sequencing data, and importantly, the associated sample and experimental metadata3. The SRA continues to grow, adding more data by the day. It is therefore an important resource for researchers who plan to integrate existing public data into their projects for meta-analysis.
The SRA enables researchers to upload and download sequencing experiments, allowing for replication, refinement, addition, and correction of any experiment involving a sequencing component. It provides a crucial touchpoint for studying experimental conditions, allowing researchers to expand their comparisons without increasing their sequencing costs. Its open access allows researchers to publish without having to shoulder the costs of storing, sending, or validating requests for data.
Data in the SRA is organized around a hierarchy4 shown below:
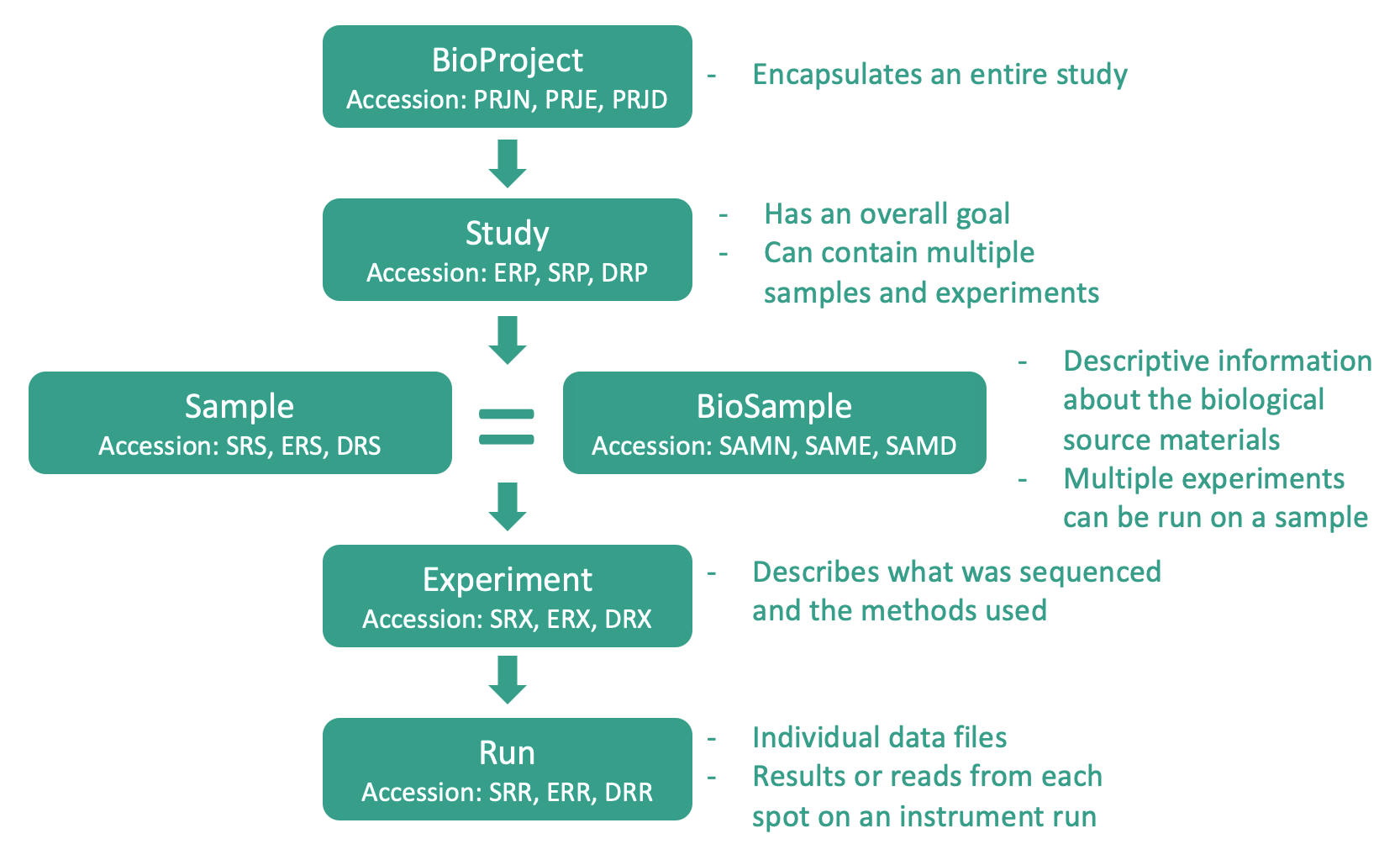
Metagenomics in the SRA
The amount of metagenomic sequence data in the SRA has increased exponentially over the years. The repository now contains over 2 million samples labeled as metagenomic. Combined with associated metadata, this enables metagenomic research to expand in its scope and makes meta-analysis easier.
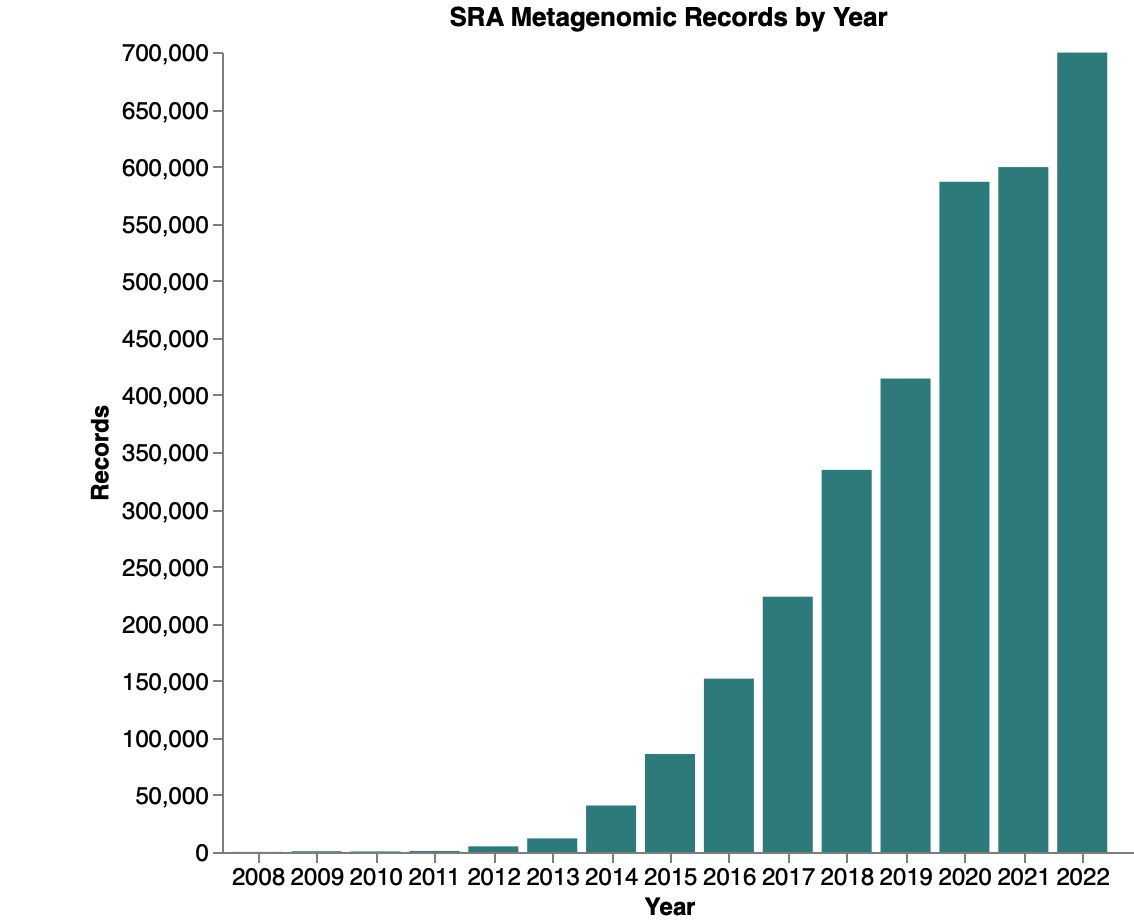
Metagenomics records in the Sequence Read Archive as of Jan. 3rd, 2023.
SRA Data in One Codex
Recognizing the importance of the resource and the needs of researchers, it is now possible to easily import SRA data into your One Codex account. We support importing all accession types in the SRA hierarchy, and imports include the metadata associated with each sample. This will make it easier for you to analyze historical data with the latest One Codex classifications and tools, and compare your samples to those of other studies of interest.
The key features that we’ve included are:
- Import a BioProject, Study, BioSample, Sample, Experiment, and Run automatically into a One Codex project
- Import data from the European Nucleotide Archive (ENA) or the DNA Data Bank of Japan (DDBJ)
- Import batches of samples without duplicating them
- Automatically include the metadata associated with each sample
- Automatically launch One Codex classification analysis upon import
Instructions for using the SRA importer can be found here.
The demo above shows how we imported 172 samples in just a few clicks using the BioProject Accession provided by Tamburini et al in their publication on urban and rural South African gut microbiomes5.
This tool allows for the quick and organized import of SRA data. One Codex continues to innovate and improve the data delivery experience for researchers to break down the barriers to project success. Stay tuned for future blogs on using SRA data in the One Codex platform!