Cataloging the Global Diversity of the Human Gut Microbiome

The microbial communities that inhabit our bodies can be thought of as unique fingerprints that both reflect and help determine many aspects of our health. Currently, metagenomic sequencing offers the best window we have into our microbiomes, which are primarily composed of uncultivated organisms. However, despite important insights gained from 20+ years of sequencing human microbiomes, it is estimated that 40-50% of species in the human gut microbiome lack reference genomes (Nayfach et al, 2019, Nature). Science is still trying to answer the question of who lives in our guts, what differences exist between people, and the underlying mechanisms behind these differences.
On a global scale, differences in gut microbiomes between human populations are often viewed through the broad lenses of urbanization and Westernization. However, samples from the United States, Europe, and China are highly overrepresented in gut microbiome data (Almeida et al, 2021, Nature Biotechnology). A notable body of work exists on microbiomes of undersampled populations, such as rural populations in Malawi (Yatsunenko et al, 2012, Nature) and Burkina Faso (Filippo et al, 2010, PNAS), Amerindians in Venezuela (Yatsunenko et al, 2012, Nature), agrarian Fiji Islanders (Brito et al., 2016, Nature), and hunter-gatherer communities (Obregon-Tito et al, 2015, Nature Communications; Schnorr et al, 2014, Nature Communications; Smits et al., 2017, Science). Analyses of these cohorts have shown that intraspecies diversity can map with geographic location and revealed entire bacterial groups abundant in non-Westernized individuals that are essentially absent in Western individuals. Functional specialization between microbiomes of different populations was also observed. However, a continuous effort to counter sampling bias and catalog the full diversity of human microbiomes is still needed.
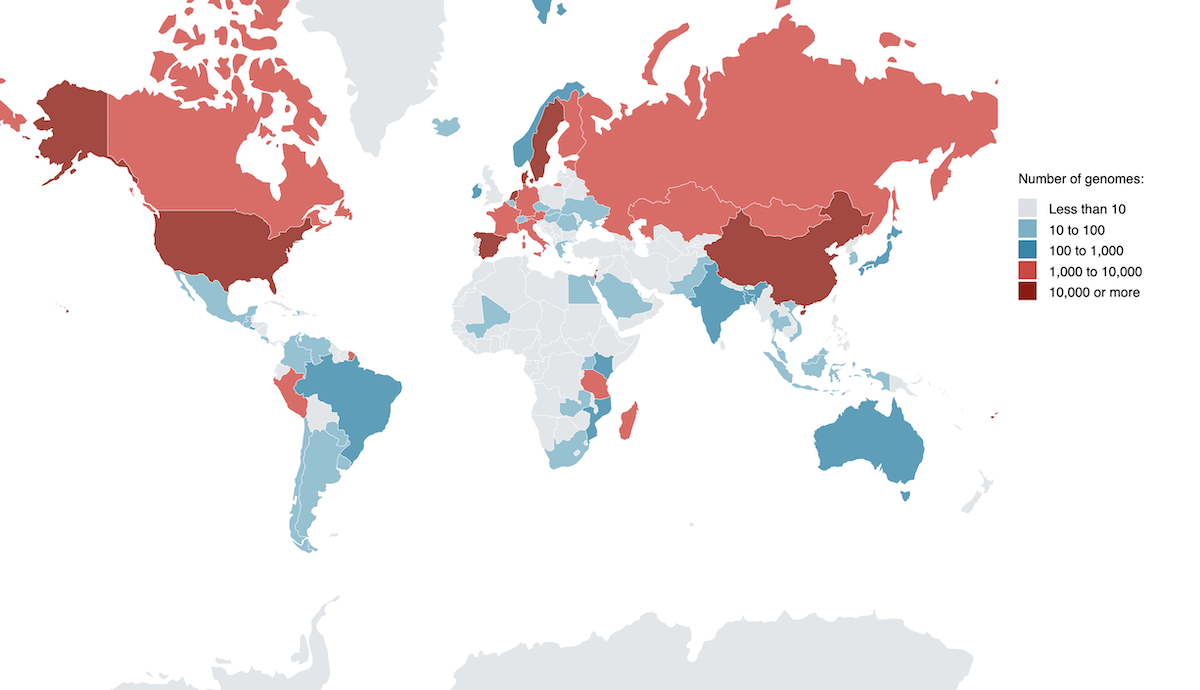
A survey of human gut microbiome reference genomes by geographic location. Metadata from Almeida et al, 2021, Nature Biotech
In tandem with continued sampling, large strides can be made by re-analyzing existing microbiome data with new metagenomic analysis techniques. In the last few years, consortia such as the European Bioinformatics Institute and the US Department of Energy Joint Genome Institute have made massive efforts to reconstruct individual microbial genomes from Tb of publicly available metagenomic data. These newly generated gut microbiome genomes–called metagenome-assembled genomes (MAGs)–can vastly improve reference genome databases, increasing the mappability of reads from Asian and African individuals by up to five-fold (Almeida et al, 2021, Nature Biotechnology). We still have a long way to go, but the ever-increasing body of microbiome sequencing data and advances in metagenomic analysis will continue to reveal unexplored diversity in the human gut microbiome.
References
- Nayfach, S., Shi, Z.J., Seshadri, R., Pollard, K.S., Kyrpides, N.C. New insights from uncultivated genomes of the global human gut microbiome. Nature, 2019. https://doi.org/10.1038/s41586-019-1058-x
- Almeida, A., Mitchell, A.L., Boland, M., Forster, S.C., Gloor, G.B., et al. A new genomic blueprint of the human gut microbiota. Nature, 2021. https://doi.org/10.1038/s41586-019-0965-1
- Yatsunenko, T., Rey, F.E., Manary, M.J., Trehan, I., Dominguez-Bello, M.G., et al. Human gut microbiome viewed across age and geography. Nature, 2012. https://doi.org/10.1038/nature11053
- Filippo, C.D., Cavalieri, D., Paola, M.D., Ramazzotti, M., Poullet, J.B., et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. PNAS, 2010. https://doi.org/10.1073/pnas.1005963107
- Brito, I.L., Gurry, T., Zhao, S., Huang, K., Young, S.K., et al. Transmission of human-associated microbiota along family and social networks. Nature Microbiology, 2019. https://doi.org/10.1038/s41564-019-0409-6
- Obregon-Tito, A.J., Tito, R.Y., Metcalf, J., Sankaranarayanan, K., Clemente, J.C., et al. Subsistence strategies in traditional societies distinguish gut microbiomes. Nature Communications, 2015. https://doi.org/10.1038/ncomms7505
- Schnorr, S.L., Candela, M., Rampelli, S., Centanni, M., Consolandi, C., et al. Gut microbiome of the Hadza hunter-gatherers. Nature Communications, 2014. https://doi.org/10.1038/ncomms4654
- Smits, S.A., Leach, J., Sonnenburg, E.D., Gonzalez, C.G., Lichtman, J.S., et al. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science, 2017. https://doi.org/10.1126/science.aan4834