COVID-19 Sequencing Analysis
SARS-CoV-2, the virus responsible for the disease COVID-19, is now widespread within our communities and has personally affected members and friends of our team.
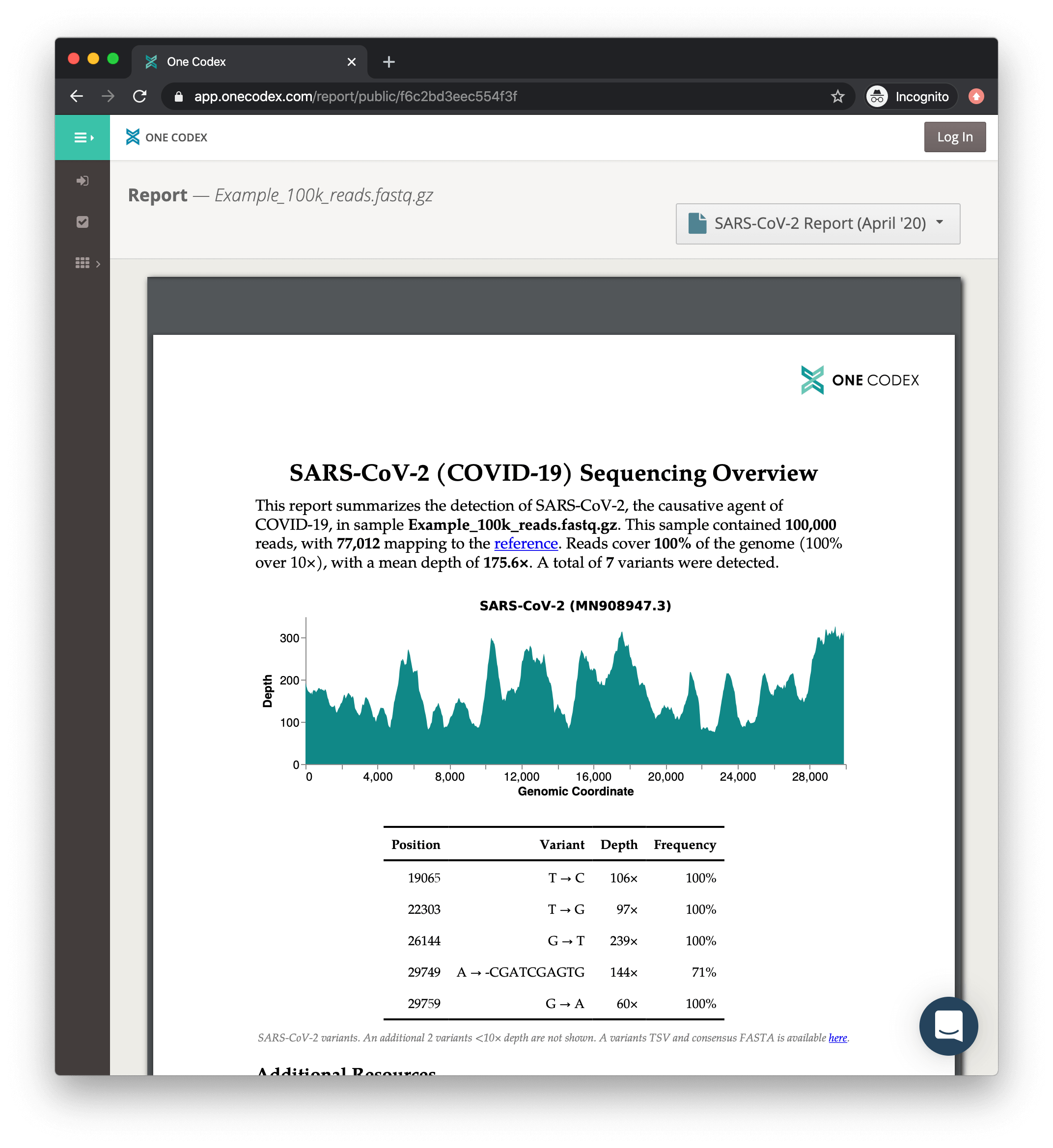
In an effort to contribute however we can, our team has been working to make analysis of genomics data from COVID-19 samples simpler, more accessible, and easier to share with the broader community. Today, we’re announcing a new analysis summarizing the detection of SARS-CoV-2 in samples that contain evidence of the virus. We’re making this analysis available at no charge and it will be automatically run on any samples in which we detect SARS-CoV-2. In return, all that we ask is that you share relevant metadata and the consensus sequences from your samples (and we’ll help enable that).
Additional details: You can view more details about the analysis in our documentation portal.
We are also working closely with current and new customers, partners, and academic collaborators to help with the response to COVID-19 wherever possible.
Our platform supports analysis of data from capture-based enrichment protocols, amplicon sequencing, and RNA sequencing (RNA-seq) of SARS-CoV-2 samples.
A Few Notes on Data Sharing and Security
We believe it’s important to follow the World Health Organization (WHO) guidance on data sharing and contribute to combat the current COVID-19 pandemic. We highly encourage everyone with SARS-CoV-2 data to share it, through our platform or otherwise. For our part, we are working to publish data through public initiatives designed specifically for outbreaks (e.g., GISAID).
One Codex is a HIPAA-compliant platform and takes safeguarding our users’ data extremely seriously, including the security of Protected Health Information (PHI). If you plan to upload PHI to One Codex or have any data security concerns, please don’t hesitate to reach out to us.
Published Protocols for Sequencing SARS-CoV-2 Samples
- Twist Bioscience has made SARS-CoV-2 RNA controls and panels for targeted enrichment
- The ARTIC Network has published an amplicon sequencing protocol for ONT’s MinION
Other Resources
- Nextstrain maintains an up-to-date analysis of SARS-CoV-2 (HCoV-19)
- The Global Initiative on Sharing All Influenza Data (GISAID) hosts viral genomes from ongoing outbreaks