2.0!
Today we’re excited to announce a major update to One Codex, which includes both improvements to our core metagenomics pipeline and an expansion of our reference database. Along with this update, we’ve also re-analyzed all samples previously uploaded to One Codex (all older analyses of course remain available).
Improved classifier: Better filtering, while maintaining sensitivity
Over the past few years, a number of new k-mer based metagenomic classifiers tools have been developed, including Kraken, GOTTCHA, CLARK, and our own. These methods have enabled ever-larger reference libraries and provided extremely sensitive detection. As a consequence of their design, however, they have also been more prone to false positives than more conservative alignment- or marker-based approaches.
We’ve previously worked to address this challenge through both substantial curation of our reference database and building secondary, confirmatory panels for specific pathogens. While we believe these efforts have made One Codex the best platform for metagenomics, there have remained use cases where – quite frankly – we and other bioinformatics tools continue to fall short.
With this newest release, we’ve made substantial strides towards eliminating false positives in complex metagenomic samples, while also adding estimated abundance and depth metrics to our results for whole-genome shotgun data. With today’s updates One Codex provides the best of both worlds – exquisite sensitivity that’s more robust to false positives.
The below figure compares the latest version of One Codex against Kraken and MetaPhlAn using an in silico simulated sample from Segata et al. (2012). It shows how the latest version of the platform provides extremely accurate relative abundance estimates (a strength of MetaPhlAn), while also substantially limiting the number of false positives when compared to previous k-mer based methods like Kraken.
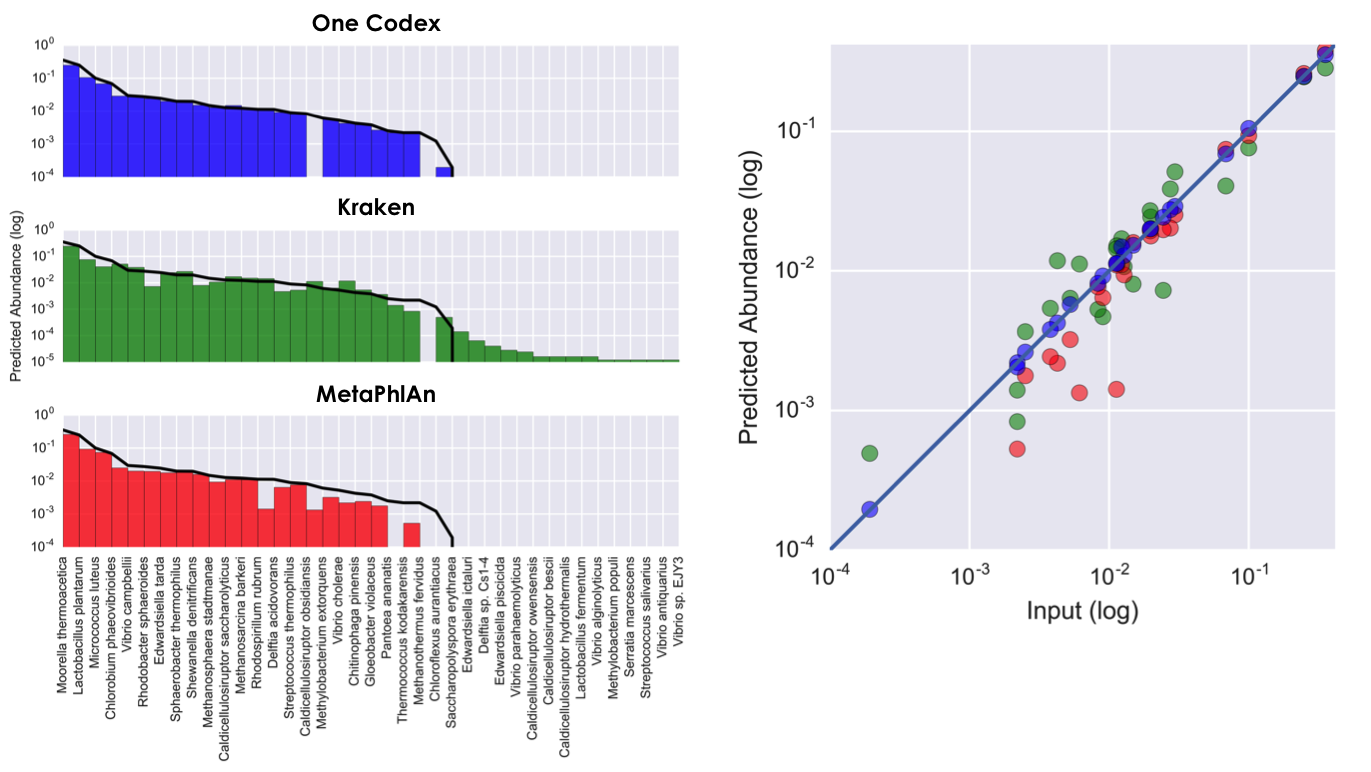
R-squared between each method and the input set are 0.9998 (One Codex), 0.9889 (Kraken), and 0.9988 (MetaPhlAn). The above figure shows 14 false positives found by Kraken at abundance >= 0.001% (an additional 112 false positives are not shown; no false positives are displayed at right). One Codex results for the dataset are available here. One Codex finds little species-specific evidence for both missing truth set species, but assigns reads to their respective genera.
Reference database updates: Spanning the tree of life
Beyond these computational updates, we’ve also further expanded our reference database, which now includes nearly 40,000 genomes across 11K distinct bacterial species, 4K viral species, and hundreds of archeal and eukaryotic species (and even more strains!). Some highlights include enhanced coverage of fungal genomes (874 total references) and protists (133), including 8 strains of Toxoplasma gondii. We’ve also added strains across a number of fungal and protozoan pathogens: this update includes 2x the number of Plasmodium falciparum and 3x the number of Trichophyton rubrum references.
Questions? Comments?
As always, please feel free to drop us a note if you have any questions, feedback, or would like to discuss a project.