New features: Whole genome clustering and BaseSpace integration
Today we’re happy to announce two new features for all users on One Codex: whole genome clustering and integration with Illumina’s BaseSpace. The first opens the door to new types of analyses, while we hope the second will allow many to spend less time moving their data around and more time exploring it!
Whole (meta)genome clustering
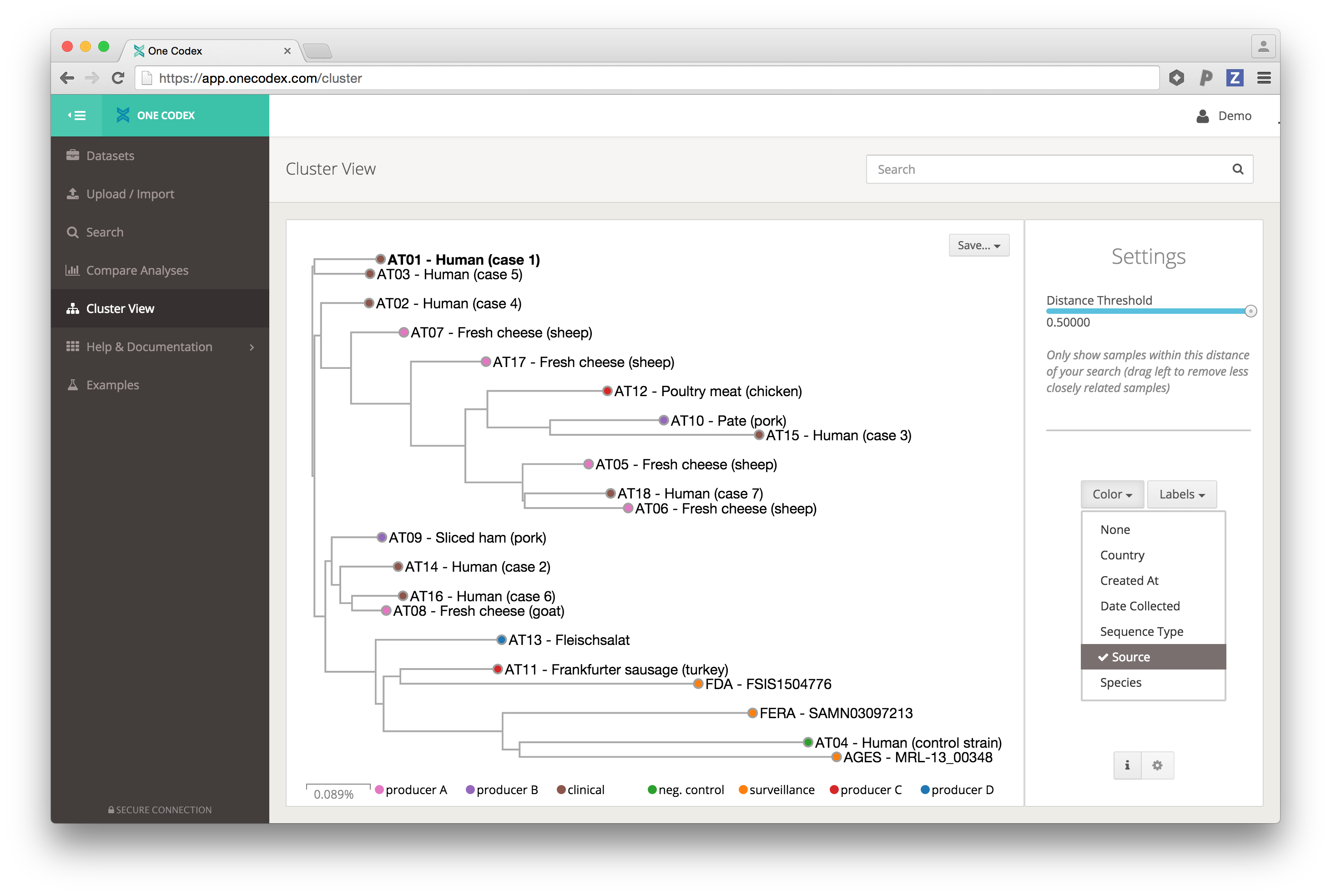
In addition to our previous sample comparison tool, we’re very excited to announce that One Codex now supports arbitrary, interactive exploration and clustering of your isolates and metagenomic samples. This cluster view (login/free registration required) enables rapid, reference-free exploration and comparison of NGS samples, often both highlighting expected similarities and revealing important inter-sample differences.
The example below shows a group of bacterial isolates from an outbreak of Listeria monocytogenes in Germany and Austria in 2011-2013 in which investigators used whole-genome sequencing to track the source to a set of food manufacturing plants. The samples group by their overall genomic similarity (not just a select set of core genome SNPs) and provide a powerful data-driven starting point for epidemiological investigation. The color of each point indicates the source it was isolated from, and it’s easy to see that the majority of clinical samples are closest to the two facilities (code-named A and B) implicated in the outbreak.
This whole genome clustering works by comparing the complete genomic content of every FASTA or FASTQ, independent of any reference database – capturing strain-level differences between isolates, and community-level differences between complex microbiome samples. (More details coming in a longer post soon!)
BaseSpace Import
We’re also happy to announce an integration with Illumina’s BaseSpace, allowing you to directly import data already in BaseSpace into One Codex. We know many will find this a useful complement to our current web- and command line client-based uploaders.
To get started importing samples from BaseSpace, simply select BaseSpace from the Upload / Import options:

After being prompted to log in to your BaseSpace account, you will be able to browse, select, and import samples from your BaseSpace projects (shown below).
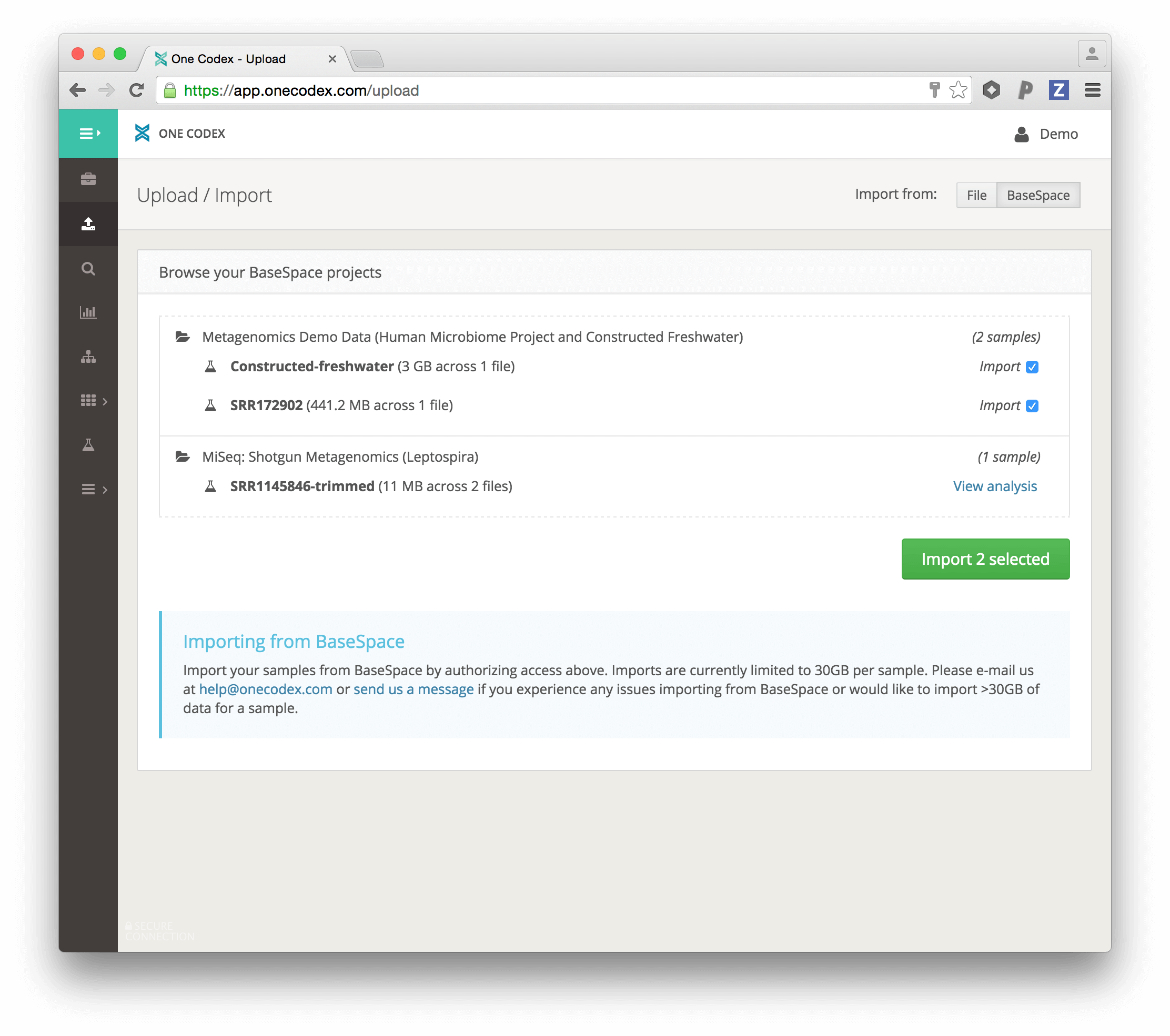
After each sample is imported, it will be automatically analyzed and available in your One Codex account. In addition, a summary report will also be created and placed in your BaseSpace account. We hope this reduces the time we all spend wrangling large NGS files, and get to analyzing our microbial samples more quickly.
Thoughts? Questions? Feedback?
As always, we’d love to hear your thoughts and feedback. And while we plan to share more details on our whole genome clustering approach in a longer post soon, please don’t hesitate to send us a note in the meantime or just stay tuned!