Automated MLST annotation
Today we added automated Multi-Locus Sequence Typing (MLST) to One Codex.
MLST is a powerful epidemiological tool that is based on curated collections of conserved mutations in core marker genes. This common reference standard is used to differentiate closely-related isolates of the same species, with many common species having hundreds or thousands of defined MLST profiles.
Datasets that are identified as being isolates or single-genome assemblies will be automatically analyzed and tagged with the detected ST label. This MLST tagging is implemented for the most commonly analyzed bacteria, including E. coli, S. enterica, and L. monocytogenes.1
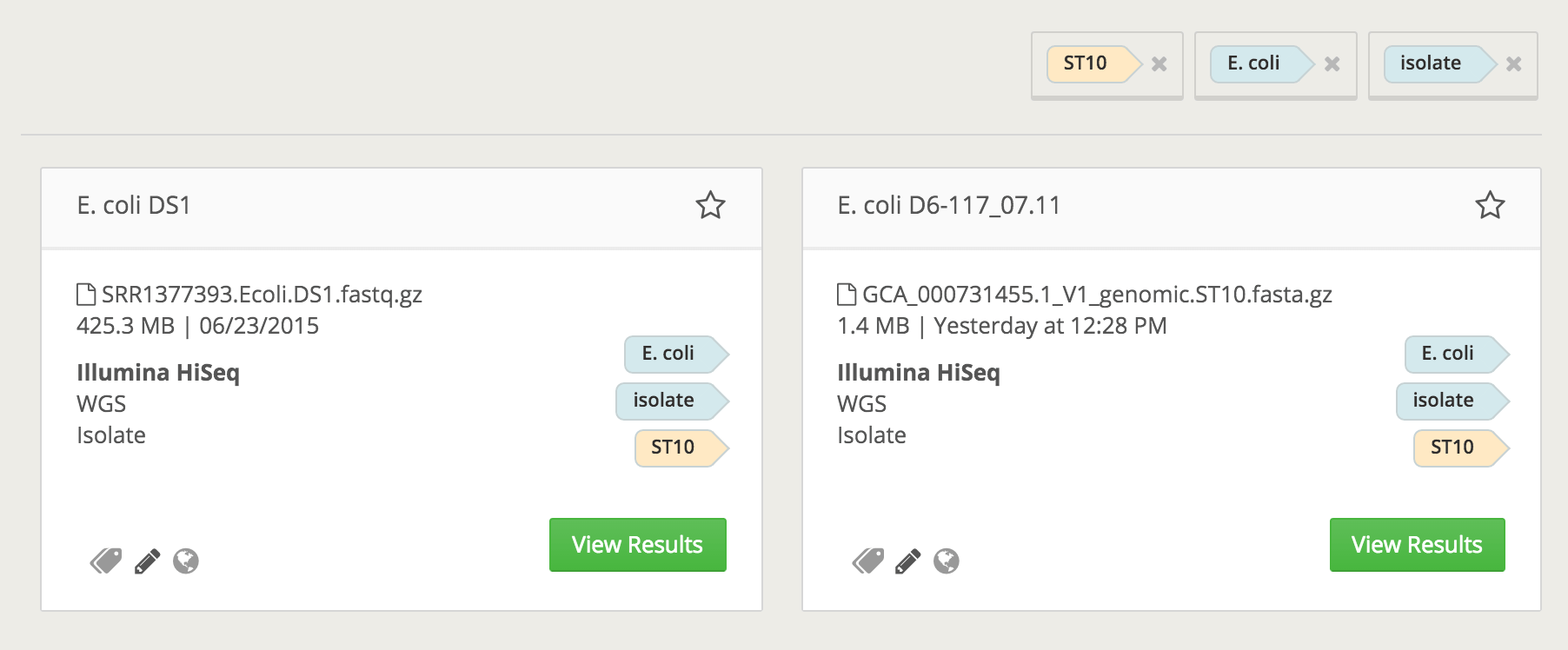
With this new feature, you can now search and filter by MLST, quickly identifying all samples with the same annotated species and sequence type (ST). This is useful in cluster and outbreak investigations, as well as other epidemiological use cases.
Please don’t hesitate to get in touch if you have any feedback, suggestions, or requests. We hope this is a useful complement to the whole genome classification on One Codex. More coming soon!
1 One Codex will also automatically add ST types to any S. aureus, S. pneumoniae, E. faecium, C. jejuni, A. baumannii, K. pneumoniae, H. influenzae, and B. pseudomallei samples using the PubMLST and University of Warwick databases. Note that we only tag samples that unambiguously match a known MLST scheme.